 |
| Home |
| Research |
| People |
| Publications |
| Books |
| Contact |
| Research Highlights |
| Group Event |
| Links |
| Research Topics in the He Group
|
Our research program spans a broad range of chemical biology, nucleic acid chemistry and biology, epigenetics, cell biology, bioinorganic chemistry, structural biology, microbiology, and genomics. We probe the pathways and mechanisms of nucleic acids modification and demodification. We study virulence and antibiotic resistance regulation in human pathogens. We also study selective metal ion recognition and sensing by naturally occurring and engineered proteins, and live-cell imaging of metal ions and other small molecules such as H2S, heme, and CO.
1. Reversible RNA Methylation: towards RNA Epigenetics
Cellular RNAs contain more than a hundred structurally distinct post-transcriptional modifications at thousands of sites. Some RNA modifications are dynamic and may have critical regulatory roles analogous to those of protein and DNA modifications. Understanding the scope and mechanisms of dynamic RNA modifications, thus, represents an emerging research frontier in biology and medicine. The internal N6-methyladenosine (m6A) modification in messenger RNA is one of the most abundant RNA modifications in eukaryotes. This base modification is present in 3-5 sites on average of every mRNA in mammals. Deletion of this ubiquitous modification leads to apoptosis in mammalian cells and arrests development of plant cells. Yet, the functional role of m6A in mRNA has never been elucidated. We have shown for the first time that m6A in mammalian mRNA is oxidatively demethylated in vitro and inside cells by FTO (fat mass and obesity-associated protein), a major obesity factor. This and other results from our laboratory indicate the presence of a new mode of biological regulation through reversible RNA methylation in mammalian cells, which we plan to establish as a new paradigm of RNA biology.

2. DNA Methylation, Hydroxymethylation, and Oxidative Demethylation
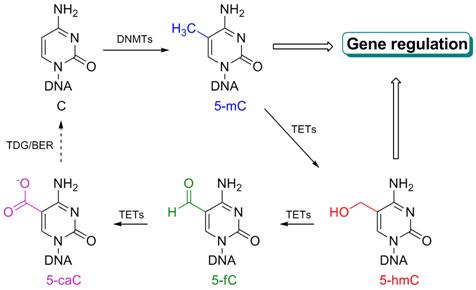
DNA is not merely a combination of four genetic nucleobases, namely, A, T, C, and G. It also contains modifications that play crucial roles throughout biology. For example, 5-methylcytosine (5-mC), the fifth DNA base which is also a crucial epigenetic mark, constitutes ~2-8% of the total cytosines in human genomic DNA and impacts a broad range of biological functions. Recently, the presence of oxidized 5-mC, 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC), and 5-carboxylcytosine (5-caC), have been discovered in mammalian cells and tissues as the sixth, seventh, and eighth DNA bases. A group of iron(II)/αKG-dependent dioxygenases, the TET proteins, have been shown to utilize dioxygen to oxidize 5-mC to these new base modifications in the mammalian genome. These discoveries strongly indicate 5-hmC as another vital epigenetic mark that plays broad roles in gene regulation, and 5-caC/5-fC as intermediates in active DNA demethylation processes. We have been working on developing effective sequencing technology to dissect the exact functional roles of these newly discovered DNA base modifications.
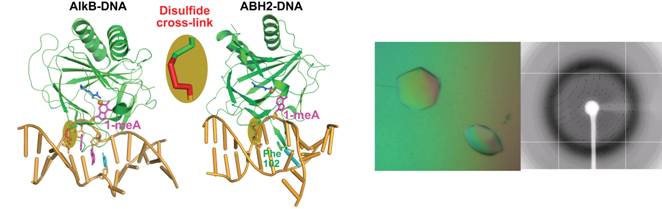
3. DNA Repair and Protein-DNA Interactions
Accumulation of genetic changes due to unrepaired DNA lesions can lead to cancer and other diseases. One component of our research program is to develop and apply a novel chemical cross-linking technique to stabilize protein-DNA interactions in distinct states in these systems. An integrative approach uniting chemical synthesis, structural biology and biochemical/biophysical characterization is used to study these interactions in DNA/RNA repair AlkB family proteins and other DNA/RNA base repair and modification proteins.
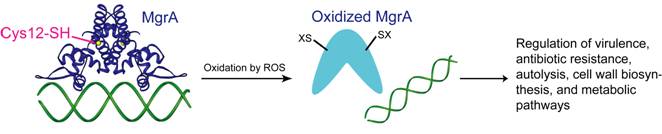
4. Virulence and Antibiotic Resistance Regulation in Human Pathogens
Staphylococcus aureus and Pseudomonas aeruginosa are human pathogens responsible for most wound and nosocomial infections. The extensive use of antibiotics to treat infections has led to the emergence of high-level resistances in various strains of these pathogens. Virulence suppression provides an alternative strategy to effectively reduce pathogenic potential without asserting selective pressure for developing drug resistances. A recent discovery in our laboratory has identified the MgrA protein as a key virulence regulator in S. aureus. This protein belongs to the MarR family of transcriptional regulators that controls antibiotic resistance and virulence in various bacteria. We demonstrated that the mgrA knockout strain shows a 10,000-fold reduction of virulence in vivo. Subsequently, we discovered that oxidative stress leads to dissociation of MgrA from its promoter DNA. The host immune response to S. aureus infection is to produce reactive oxygen and nitrogen species to counter the pathogen. Our study suggests that the microorganism uses MgrA and related regulatory proteins to sense the oxidative stress response generated by the host and regulate a global defensive response. We plan to fully elucidate the underlying virulence regulation pathways, and exploring strategies to suppress S. aureus virulence by targeting virulence regulation. We are also studying MgrA homologues in S. aureus, P. aeruginosa and other pathogens. Our ultimate goal is to develop new strategies for treating infections.
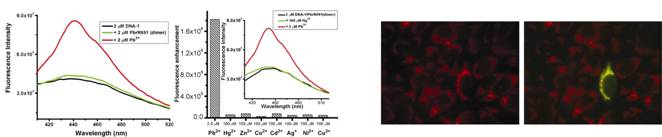
5. Selective Metal Ion Recognition by Proteins
The ability to regulate essential or toxic metal ion concentrations is critical for cell survival. Our goal is to understand how specific metal ions are recognized and regulated in biological systems. We have been working on elucidating the mechanisms of proteins that exhibit remarkable selectivity toward metal ions such as lead(II), cadmium(II), gold(I), copper(I) and iron(II). Some of these proteins can be converted into genetically encoded fluorescent probes for sub-cellular metal ion imaging in live cells. We also work on engineering proteins that possess high sensitivity and selectivity toward various metal ions including actinides, and developing probes for imaging of cellular small molecules.
Selected References
Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Yang, Y.-G. and He, C.* “N6-methyladesosine is nuclear RNA is a major substrate of the obesity-associated FTO” Nature Chem. Biol. 2011, on-line.
Qian, Y.; Karpus, J.; Kabil, O.; Zhang, S.-Y.; Zhu, H.-L.; Banerjee, R.; Zhao, J.; He, C.* “Selective
fluorescent probes for live-cell monitoring of sulfide” Nature Commun. 2011, on-line,
He, Y.; Li, B.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; Sun, Y.; Li, X.; Dai, Q.; Song, C.-X.; Zhang, K.; He, C.; Xu, G.* “Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA” Science, 2011, 333, 1303-1307.
Ito, S.; Shen, L.; Dai, Q.; Wu, S. C.; Collins, L. B.; Swenberg, J. A.; He, C. and Zhang, Y.* “Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine” Science, 2011, 333, 1300-1303.
Song, C.-X.; Szulwach, K. E.; Fu, Y.; Dai, Q.; Yi, C.; Li, X.; Chen, C.-H.; Zhang, W.; Jian, X.; Wang, J.; Zhang, L.; Looney, T. J.; Zhang, B.; Godley, L. A.; Hicks, L. M.; Lahn, B. T.; Jin, P.* and He, C*. “Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine” Nature Biotechnol., 2011, 29, 68-72.
Yi, C.; Jia, G; Hou, G.; Dai, Q.; Zhang, W.; Zheng, G.; Jian, X.; Yang, C.-G.; Cui, Q.; He, C.* “Iron-catalysed Oxidation Intermediates Captured in a DNA Repair Dioxygenase” Nature, 2010, 468, 330-333.
Yang, C.-G.; Yi, C.; Duguid, E. M.; Sullivan, C. T.; Jian, X.; Rice, P. A. and He, C.* “Crystal Structures of DNA-RNA Repair Enzymes AlkB and ABH2 Bound to dsDNA” Nature, 2008, 452, 961-965.
Chen, P.; Bae, T.; Williams, W. A.; Duguid, E. M.; Rice, P. A.; Schneewind, O. and He, C.* “An Oxidation Sensing Mechanism is Used by A Global Regulator of Staphylococcus aureus” Nature Chem. Biol. 2006, 2, 591-595.
Chen, H.; Hu, J.; Chen, P. R.; Lan, L.; Li, Z.; Hicks, L. M.; Dinner, A. R. and He, C.* “The Pseudomonas aeruginosa Multidrug Efflux Regulator MexR Uses An Oxidation Sensing Mechanism” Proc. Natl. Acad. Sci. 2008, 105, 13586-13591.