Accumulation of genetic changes due to the presence of
unrepaired DNA lesions can lead to cancer development and other diseases.
One component of our research program is to develop and apply a novel
chemical cross-linking technique to stabilize protein/DNA interactions in
distinct states in these systems. An integrative approach uniting chemical
synthesis, structural biology and biochemical characterization is used to
study these interactions in DNA repair AGT and AlkB proteins and other DNA
base repair and modification proteins. In the other component of the
research, we seek to develop a chemical proteomic approach to trap and
identify undiscovered base repair and modification proteins. By preparing
various DNA probes we plan to covalently trap proteins that specifically
interact with these probes. New proteins or new functions could be
identified.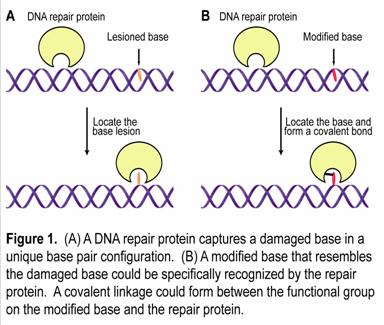
The basic premise of our approach is that every damage repair protein must possess a mechanism to search for the damaged bases. We envision that a repair protein may capture a damaged base by recognizing a unique base pair configuration (Figure 1A); for instance, a damaged base could give rise to an unstable base pair. Then, we can chemically synthesize a modified base bearing certain functional groups. The base can be incorporated into a base pair that resembles the property of the damaged base pair. The repair protein may specifically recognize this modified base and insert it into the active site of the protein (Figure 1B). The functional group on the modified base was designed to be able to crosslink to the active site residues of the repair protein. Therefore, a covalently trapped protein/DNA complex can be prepared for structural studies or for identification of the protein directly from cell extracts. Since the method relies on the intrinsic property of the repair protein to search for corresponding base lesions, the cross-linking reaction is driven by the protein/DNA interaction and should be efficient compared to random, nonspecific reactions.
a) O6-alkylguanine-DNA alkyltransferases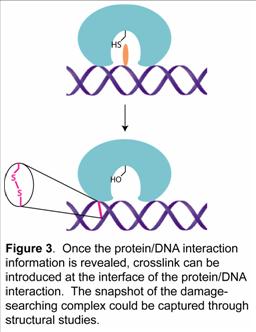
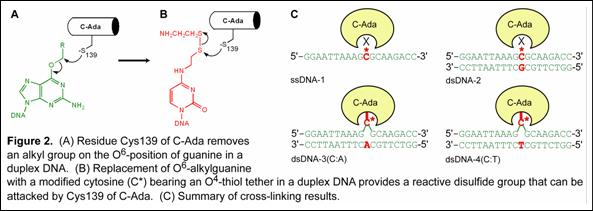
We have utilized the strategy and successfully crosslinked bacterium and human O6-alkylguanine-DNA alkyltransferases with modified DNA (Figure 2). The structural and functional studies of these proteins are in progress in our group. Once the detailed protein/DNA interaction is revealed, we will apply the chemical cross-linking method to trap the nonspecific protein/DNA complexes (Figure 3). Structural study of this complex would reveal the molecular detail of the damage-searching mechanism of DNA repair proteins.
b) AlkB
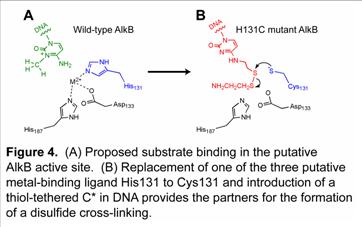
The AlkB proteins is a newly discovered DNA base repair protein family that has attracted great attention due to its unique repair activity. Several human homologues have also been found. We plan to elucidate the mechanism and the structure of the AlkB proteins. Native iron(II)-containing AlkB was isolated in out laboratory. We plan to prepare modified oligonucleotides to probe its mechanism and apply spectroscopic methods to study the iron(II) center. We also apply the chemical cross-linking technique to prepare and isolate AlkB/DNA complexes for structural characterization; the AlkB proteins bind DNA weakly and could not for specific complexes without covalent cross-linking.